Isolation and Screening of Antibiotic producing Bacteria from Soil in Lahore City
1Department of Zoology, Lahore College for Women University, Jail Road Lahore 54000, Pakistan.
2The School of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
3Pakistan Science Mission (PSM), Noor Kot 51770, Pakistan.
4Yunnan Institute of Microbiology, Yunnan University, Kunming, 650091, PR China.
*Corresponding author:
Muhammad Naeem Iqbal;Email:
driqbalnaeem@hotmail.comViews 3438
Abstract
The need for new antibiotics is increasing with the increase in antibiotic resistance. The present research work has been focused on the isolation of antibiotic producing bacteria from the soil samples collected from Jinnah garden, Lahore. The isolates were identified based on their morphology and further confirmed through biochemical tests. Antibiotic producing ability was confirmed by inhibition zones around bacterial colonies. A total of 56 isolates belonging to four different strains of antibiotic producing bacteria were found in different soil samples. Out of these, 30 were Bacillus subtilis, 9 were Bacillus licheniformis, 12 were Streptomyces and 5 were Actinomycetes. A significant number of antibiotic producing bacteria were found in this study. These results suggest that soil isolates, having antibiotic producing capability can be used commercially after proper standardization.
Keywords
Screening, Isolation, antibiotics production, bacteria.
Citation
Yunus, F.N., Khalid, Z.Z., Rashid, F., Ashraf, A., Iqbal, M.N., Hussain, F., 2016. Isolation and Screening of Antibiotic producing Bacteria from Soil in Lahore City. PSM Microbiol., 01(1): 01-04.
Introduction
Antibiotics can be defined as any chemical substance of small organic molecules which at low amounts are harmful to the metabolic activities and growth of other microbes (Brun and Skimkets, 2000). The natural products obtained from microorganisms are still appearing as the most auspicious source of the future antibiotics (Pelaez, 2006). Antibiotics which are mostly in use today are the natural derivatives of fungi and actinomycetes (Tawiah et al., 2012). In nature, there is universal dissemination of antibiotics among the microorganisms responsible for their antagonism (Euanorasetr et al., 2010).
There are numbers of bacteria which are capable of producing antibiotics which include Bacillus (Waites et al., 2008), Actinomycetes (Abdulkadir and Waliyu, 2012; Tiwari and Gupta, 2013) Pseudomonas (Cartwright et al., 1995), and Streptomyces (Willey et al., 2008) and most of these isolates are soil bacteria.
Extensive use of antibiotics both for clinical and veterinary purposes leads to the development of resistance in many infectious strains (Kurtboke et al., 1992). With the continuation of this process of resistance, the search for new anti-infective drugs should be carried out (Thakur et al., 2007; Brown and Wright, 2005). In addition to the resistance problem, a number of new infectious diseases have been discovered over the past 30 years (Zimmerman and Zimmerman, 2003; Bryskier, 2005).
Soil is rich in microorganisms which are capable of producing antibiotics (Brun and Skimkets, 2000). The traditional approach is ‘random screening’ in which bacteria are isolated, grown and their activity spectrum was assessed. Even this has been done for more than 50 years still we are getting results in favor to us and thus we are sticking with this approach (Wawrik et al., 2007).
The aim of this study was isolation and screening of antibiotic producer bacteria from soil samples from the Jinnah garden, Lahore.
Materials and Methods
Collection of samples
A total number of 25 soil samples weighing approximately 10 gram each were collected from different areas of Jinnah garden, Lahore. All the samples were collected 4 to 5 cm deep from the surface using spatula and placed in sterilized zip lock polythene bags. The samples were transferred to Zoology Research Laboratory, Lahore College for Women University and kept at 4ᵒC for further processing.
Processing of samples
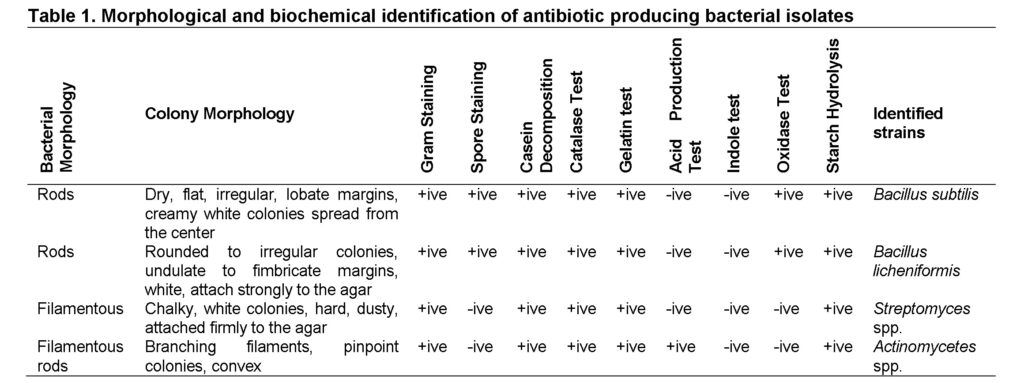
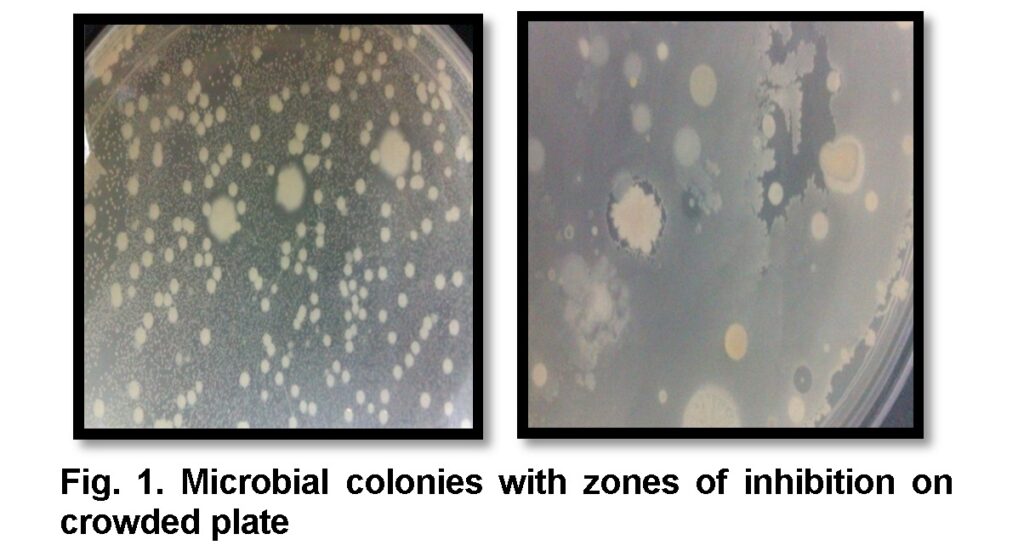
Samples were processed by dissolving 1 g of soil in 10 ml of sterile distilled water to make soil suspension in test tube. The test tube was vortexed for 2 to 3 minutes to remove soil, stones and debris. Supernatant was transferred to another test tube and ten-fold serial dilutions were prepared. Then spread 100µl of supernatant from each dilution on nutrient agar plates and kept at 37ᵒC overnight. After 24 to 48 hrs, zone of inhibition in each plate was observed and colonies were selected for further confirmation of isolates.
Isolation and Screening of bacteria
Selected colonies based on morphology and zone of inhibition were plated directly onto nutrient agar for isolation of bacteria. After an overnight incubation, discrete colonies of bacteria were selected based on colony characters and streaked on new agar plates according to previous study (Iqbal et al., 2015). The plates were then kept at 37°C for 24 hrs.
Multiple streak plate method as used previously (Iqbal et al., 2015) was applied to obtain purified cultures of bacteria on nutrient agar plates. Pure single colonies were noticed after 24 hrs incubation at 37°C.
Purified cultures of bacteria were identified based on colony morphology, microscopic characters and biochemical characters (Iqbal et al., 2015; Yunus et al., 2016; Yunus et al., 2016) following Bergey’s Manual of Determinative Bacteriology (Bergey, 1984).
Statistical Analysis
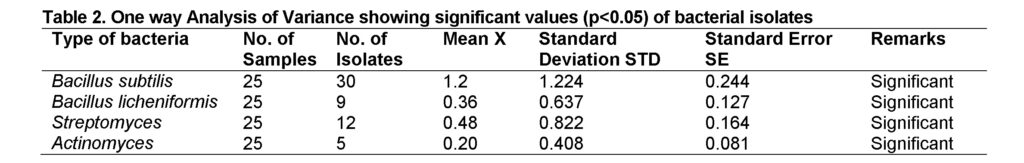
Data obtained was arranged using Microsoft Excel (Microsoft Corporation). Statistical difference between means was determined by ANOVA using SPSS version 16.0 at p˂0.05 significance level.
Results
A total of 56 bacterial fifty six isolates having zones of inhibition on crowded plate have been isolated and identified by morphological characteristics and biochemical tests from 25 soil samples (Table 1). Four different bacterial strains were present, which were Bacillus subtilis, Bacillus licheniformis, Streptomyces and Actinomycetes species. Out of these 56 isolates 30 were Bacillus subtilis, 9 were Bacillus licheniformis, 12 were Streptomyces and 5 were Actinomycetes (Table 1). Production of antimicrobial compounds was confirmed by inhibition of other bacterial growth around the colonies (Figure 1).
One way ANOVA was applied to compare means of various isolates of antibiotic producing bacteria. The bacterial isolates showed significant values (p<0.05) in soil samples (Table 2).
Discussion
Soil samples are commonly evaluated for isolation of the antibiotic producing organisms, because soil microorganisms produce lots of antibiotics in order to survive in such a competitive environment. Bacteria producing high number of medically and agriculturally important antibiotics belong to genera Bacillus, Streptomyces and Pseudomonas (Yoshiko et al., 1998; Sharga et al., 2004).
As the 39 isolates from 56 were Bacillus species and 9 isolates were Bacillus licheniformis, these results correlates with many previous studies that Bacillus spp are notorious producing antibiotics. It has been documented that Bacillus genus and other spore forming bacteria carry genes for the breakdown of diverse carbon source and production of antibiotics (Prescott et al., 2008). Bacillus species are ubiquitous in nature. Many stains of the genus Bacillus have capability to synthesize a wide range of antibiotics. Hassan et al. (2014) reported 14 isolates of antibiotic producing Bacillus species from soil samples.
Several hundred Bacillus subtilis strains have been described with the ability to synthesize more than two dozen antibiotics with diverse structures. The potential of Bacillus subtilis to synthesize antibiotics has been perceived for 50 years. The large number of antibiotics produced from Bacillus subtilis might reflect the diversity of natural isolates. Other Bacillus species like Bacillus amyloliquefaciens (Koumoutsi et al., 2004), Bacillus brevis produce few antibiotics compared to Bacillus subtilis.
Few antibiotics producing Actinomycetes have been isolated in the present work. The reason why only small number of Actinomycetes has been isolated may be the textures of soil and other predominant environmental conditions at the study location. Actinomycetes require a longer time to grow compared to other bacteria as it has been documented by previous studies (Ahmed et al., 2013). Actinomycetes can use a variety of organic nutrients but special media are often preferable (Rahman et al., 2000; Sultan et al., 2002). Probably this could be the logic why Actinomycetes grow in very small number on the media (Nutrient agar) used. Tryptic soy agar or Starch-Casein agar medium are mostly used for the isolation of Actinomycetes in many literatures.
Streptomyces are the most studied and well known group of Actinomycetes. These are gram positive, filamentous bacteria which have a great capability to synthesize most important secondary metabolites such as antibiotics, antitumor, antivirals and antifungals (Dehnad et al., 2010). Out of 56 isolates, 12 were Streptomyces having white, wrinkled colonies. Although, various biochemical tests were performed but the strains were not identified to the species level. For proper identification of genera and species of Actinomycetes as well as morphophological and physiological characteristics, different other biochemical parameters such as cell wall chemo type, peptidoglycan type, whole cell sugar pattern, phospholipid type and G+C% of DNA should be considered (Dehnad et al., 2010).
Conclusion
It is concluded that Bacillus subtilis, Bacillus licheniformis, Streptomyces and Actinomycetes isolated from Jinnah garden, Lahore have potential to produce antibiotics.
Acknowledgement
The authors are highly thankful to Dr. Fakhar-un-Nisa Yunus, Department of Zoology, Lahore College for Women University, Lahore, for technical support during this research work.
Conflict of Interest
The authors confirm that this article content has no conflict of interest.
References
Abdulkadir, M., Waliyu, S., 2012. Screening and Isolation of the Soil Bacteria for Ability to Produce Antibiotics. Eur. J. Appl. Sci., 4(5): 211-215.
Ahmed, R.N., Sani, A.H., Ajijolakewu., Alamu, F.B., 2013. Soil Screening for Antibiotic – Producing Microorganisms. Adv. Environ. Biol., 7(1): 7-11.
Bergey, S.A., 1984. Bergey, Manual of Determinative Bacteriology, 9th edition, Williams & Wilkins., Philadelphia.
Brown, E.D., Wright, G.D., 2005. New Targets and Screening Approaches in Antimicrobial Drug Discovery. Chem. Rev., 105: 759-74.
Brun, Y.V., Skimkets, L.J., 2000. Prokaryotic development. ASM Press, Pp. 11-31.
Bryskier, A., 2005. In Pursuit of New Antibiotics. In: Antimicrobial Agents. ASM Press, Washington, D.C. Pp. 1242-59.
Cartwright, D.K., Chilton, W.S. Benson, D.M., 1995. Pyrrolnitrin and phenazine produce by Pseudomonas cepacia, strain 5.5 B, a biological agent of Rhizoctonia solani. Appl. Microbiol. Biotechnol., 43: 211-121.
Dehnad, A.R., Yeganeh, L.P., Bakhshi, R., Mokhtarzadeh, A., Soofiani, S.A., Monadi, A.R., Gasanova, S., Abusov, R., 2010. Investigation antibacterial activity of Streptomycetes isolates from soil samples, West of Iran. Afr. J. Microbiol. Res., 4(16): 1685-1693.
Hassan, S.A., Hanif, E., Zohra, R.R., 2014. Isolation and screening of soil bacteria for potential antimicrobial activity. FUUAST J. BIOL., 4(2): 217-219.
Iqbal, M.N., Anjum, A.A., Ali, M.A., Hussain, F., ALI, S., Muhammad, A., Irfan, M., Ahmad, A., Irfan, M. and Shabbir, A., 2015. Assessment of microbial load of un-pasteurized fruit juices and in vitro antibacterial potential of honey against bacterial isolates. Open Microbiol. J., 9: 26-32. DOI: 10.2174/1874285820150601E001.
Koumoutsi, A., Chen, X.H., Henne, A., Liesegang, H., Hitzeroth, G., Franke, P., Vater, J., Borriss, R., 2004. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol., 186(4):1084-96.
Kurtboke, D.T., Chen, C.F., William, S.T., 1992. Use of polyvalent phage for reduction of Streptomyces on soil dilution plates. J. Appl. Bacteriol., 72: 103.
Pelaez, F., 2006. The historical delivery of antibiotics from microbial natural products_Can history repeat? Biochem Pharmacol., 71(7): 981-990.
Prescott, M.L., Harley, P.J., Klein, A.D., 2008. Microbiology. 7th Ed. Publishing Group, Pp. 42-51, 232-233, 762-764.
Rahman, M.Z.A., Azam, A.T.M.Z., Gafur, M.A., 2000. In vitro antibacterial principles of two flavonoids and extracts from Clerodendrum indicum linn. Pak. J. Biol. Sci., 3(10): 1769-1771.
Sharga, B.M., Nikolaychuk, V.I., Maga, M.I., 2004. Comparative IR Spectrometry of antimicrobial substances derived antibiotic from Bacillus. Mikpobiojiotir., 15: 75-77.
Sultan, M.Z., Khatune, N.A., Sathi, Z.S., Bhuiyan, M.S.A., Sadik, M.G., Choudury, M.A., Gafur, M.A., Rahman, M.A.A.,2002. In vitro antibacterial activity of an active metabolite isolated from Streptomyces species. Biotechnol., 1(2-4): 100-106.
Tawiah, A.A., Gbedema, S.Y., Adu, F., Boamah, V.E. and Annan, K. (2012). Antibiotic producing microorganisms from River Wiwi, Lake Bosomtwe and the Gulf of Guinea at Doakor Sea Beach, Ghana. BMC Microbiology, 12: 234-241.
Thakur, D., Yadav, A., Gogoi, B.K., Bora, T.C., 2007. Isolation and screening of Streptomces in soil of protected forest areas from the state of assam and tripura India for antimicrobial metabolites. J. Mycol Med., 17: 242-49.
Tiwari, K., Gupta, R.K., 2013. Diversity and isolation of rare actinomycetes: an overview. Crit. Rev. Microbiol., 39(3): 256-294.
Waites, M.J., Morgan, N.L., Rockey, J.S., Higton, M., 2008. Industrial Microbiology-an Introduction. Blackwell Publisher, London.
Wawrik, B., Kutliev, D., Abdivasievna, U.A., Kukor, J.J., Zylstra, G.J. Kerkhof, L., 2007. Biogeography of Actinomycetes communites and type II polyketide synthase genes in soils collected in New Jersey and Central Asia. Appl. Environ. Microbiol., 73: 2982-9.
Willey, H.J., Linda, P.S., Shewood, M., 2008. Microbiology. 5th Ed. McGraw Hill Publisher, London. P. 1035.
Yoshiko, H., Okamoto, S., Muramatsu, H.. Ochi, K., 1998. Acquisition of certain Streptomycin resistant Mutations Enhances antibiotic production in bacteria. Antimicrob Agents Chemother., 42(8): 2041-2047.
Yunus, F.N., Kanwal, F., Rashid, F., Ashraf, A., Iqbal, M.N., Xiao, S., 2016. A Comparative Study on Isolation and Identification of Bacillus thuringiensis from Different Localities of Gujranwala City. PSM Biol. Res.,01(1): 34-38.
Yunus, F.N., Iqbal, M., Jabeen, K., Kanwal, Z., Rashid, F., 2016. Antagonistic activity of Pseudomonas fluorescens against fungal plant pathogen Aspergillus niger. Sci. Lett., 4(1): 66-70.
Zimmerman, B.E., Zimmerman, D.J., 2003. Microbes and Diseases That Threaten Humanity. Contemporary Books. Chicago: New York, San Francisco.





